|
Mesenchymal stromal cells (MSCs) are multipotent cells that have great potential for regenerative medicine, tissue repair, and immunotherapy.
Unfortunately, the outcomes of MSC-based research and therapies can be highly inconsistent and difficult to reproduce, largely due to the
inherently significant heterogeneity in MSCs, which has not been well investigated. To quantify cell heterogeneity, a standard approach is
to measure marker expression on the protein level via immunochemistry assays. Performing such measurements non-invasively and at scale has
remained challenging as conventional methods such as flow cytometry and immunofluorescence microscopy typically require cell fixation and
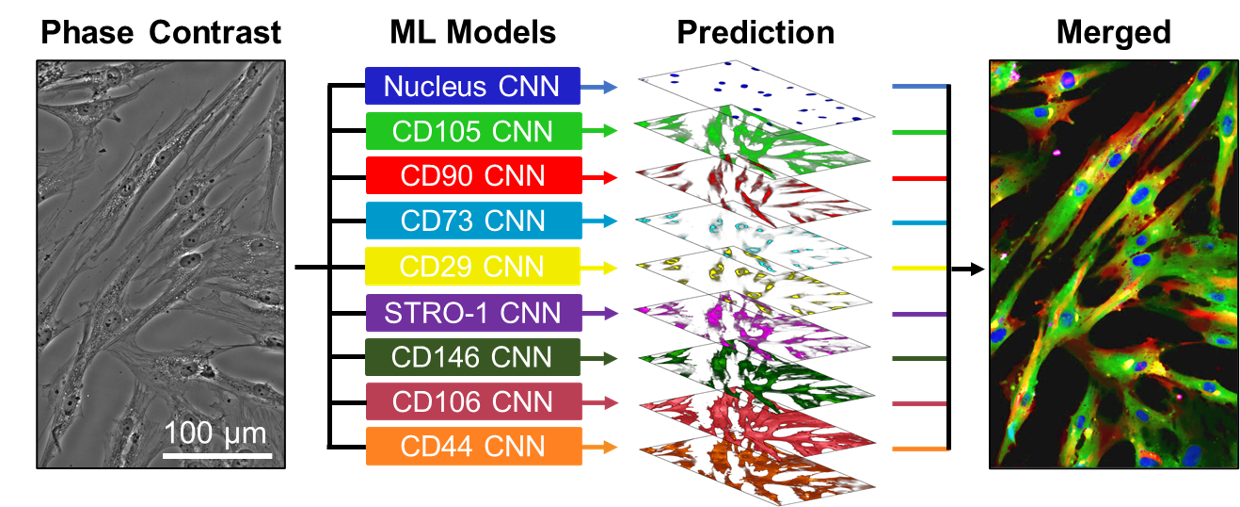
laborious sample preparation. Here, we developed an artificial intelligence (AI)-based method that converts transmitted light microscopy images
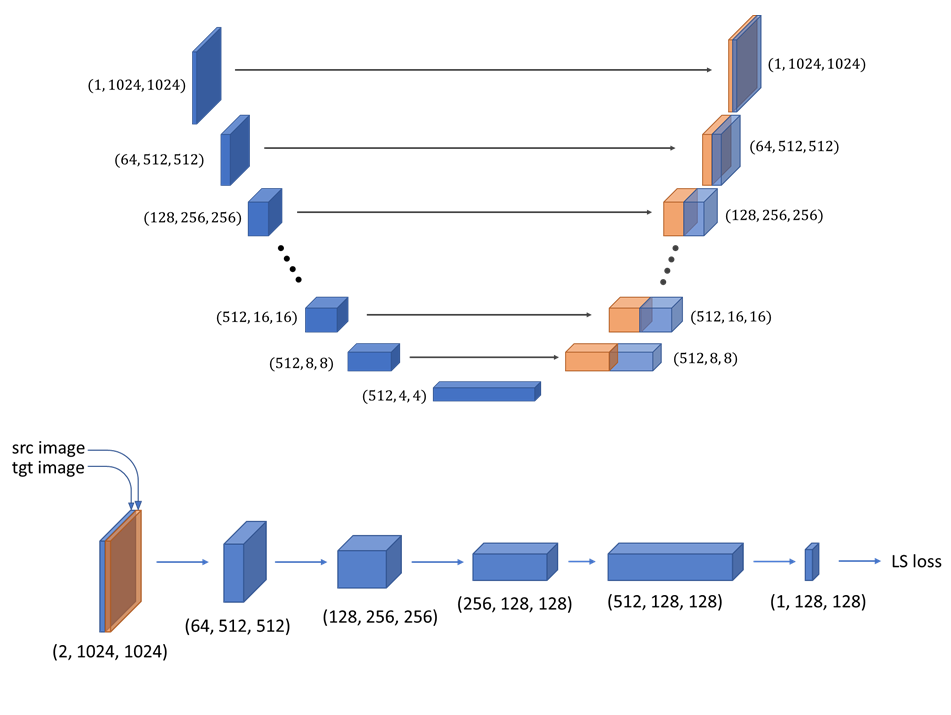
of MSCs into quantitative measurements of protein expression levels. By training a U-Net + conditional generative adversarial network(cGAN)
model that accurately (mean rs= 0.77) predicts expression of 8 MSC-specific markers, we showed that expression of surface markers provides a
heterogeneity characterization that is complementary to conventional cell-level morphological analyses. Using this label-free imaging method,
we also observed a multi-marker temporal-spatial fluctuation of protein distributions in live MSCs. These demonstrations suggest that our
AI-based microscopy can be utilized to perform quantitative, non-invasive, single-cell, and multi-marker characterizations of heterogeneous
live MSC culture. Our method provides a foundational step toward the instant integrative assessment of MSC properties, which is critical for
high-throughput screening and quality control in cellular therapies.
|